Note
Click here to download the full example code
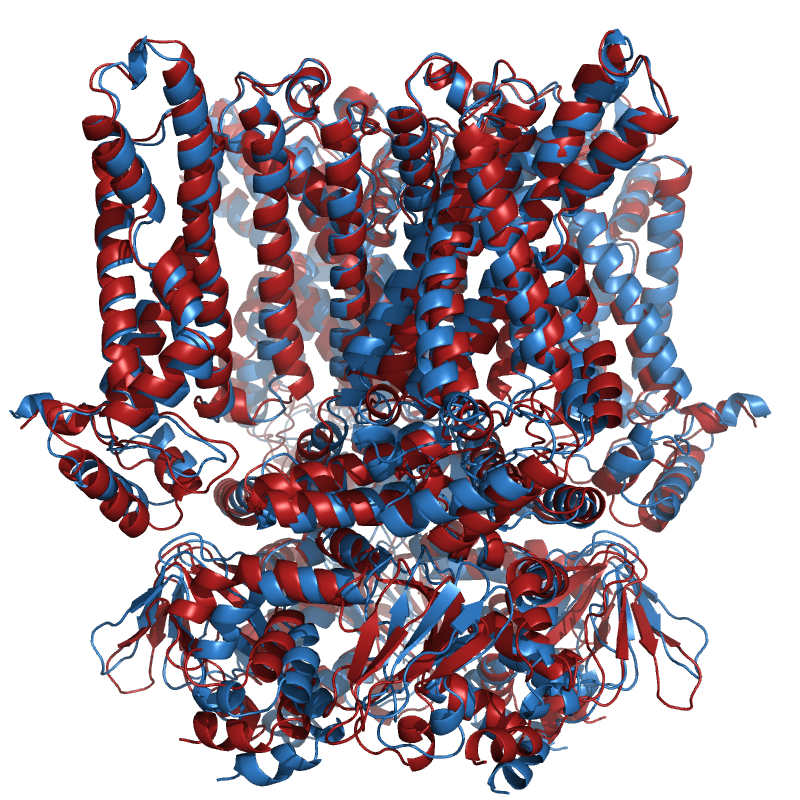
Displacement between HCN4 C-linker in apo and holo conformation¶
This example shows the extension of the C-linker ‘disc’ of the HCN4 ion channel upon binding of the ligand cAMP. The displacement between the apo and holo structure is clarified using arrows.
# Code source: Patrick Kunzmann
# License: CC0
import numpy as np
import biotite.structure as struc
import biotite.structure.io.mmtf as mmtf
import biotite.database.rcsb as rcsb
import ammolite
PNG_SIZE = (800, 800)
CLINKER_RANGE = (522, 564+1)
EXTENSION_FACTOR = 7
apo_structure, holo_structure = (
mmtf.get_structure(
mmtf.MMTFFile.read(rcsb.fetch(pdb_id, "mmtf")),
model=1, include_bonds=True
)
for pdb_id in ("7NP3", "7NP4")
)
# Holo structure is slightly longer at both ends than apo structure
# -> select common atoms
holo_structure = holo_structure[
np.isin(holo_structure.res_id, apo_structure.res_id)
]
holo_structure, _ = struc.superimpose(apo_structure, holo_structure)
# Display entire ion channel
pymol_apo = ammolite.PyMOLObject.from_structure(apo_structure)
pymol_holo = ammolite.PyMOLObject.from_structure(holo_structure)
pymol_apo.color("skyblue")
pymol_holo.color("firebrick")
pymol_apo.orient()
ammolite.cmd.turn("z", -90)
ammolite.show(PNG_SIZE)
# Show only C_linker
clinker_mask = np.isin(apo_structure.res_id, np.arange(*CLINKER_RANGE))
pymol_apo.hide("cartoon", ~clinker_mask)
pymol_holo.hide("cartoon", ~clinker_mask)
displacement = holo_structure.coord - apo_structure.coord
# Make the arrows longer than the actual displacement
# to clarify the disc extension
displacement *= EXTENSION_FACTOR
start_coord = apo_structure.coord
end_coord = apo_structure.coord + displacement
ca_mask = apo_structure.atom_name == "CA"
ammolite.draw_arrows(
start_coord[clinker_mask & ca_mask],
end_coord[clinker_mask & ca_mask],
head_radius=0.5,
head_length=0.8
)
pymol_apo.orient(clinker_mask)
pymol_apo.zoom(clinker_mask, buffer=10)
ammolite.show(PNG_SIZE)
# sphinx_gallery_thumbnail_number = 2

