Note
Click here to download the full example code
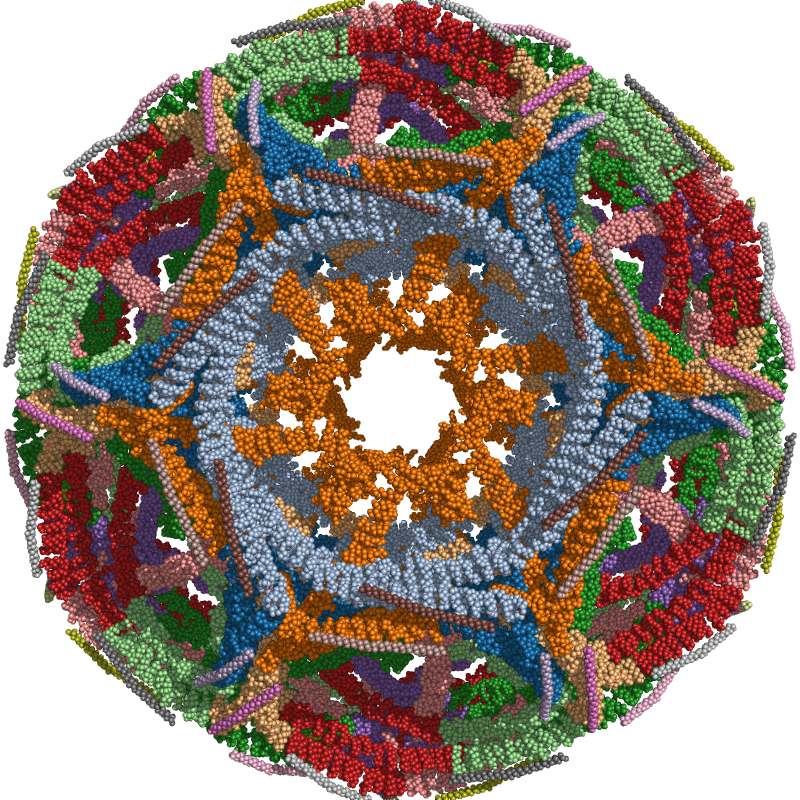
Assembly of a clathrin D6 coat¶
In this example the entire biological assembly of a clathrin D6 coat is loaded from a mmCIF file in visualized in PyMOL. Each chain is colored individually based on a qualitative Matplotlib colormap.
# Code source: Patrick Kunzmann
# License: CC0
import numpy as np
import matplotlib.pyplot as plt
import biotite.structure as struc
import biotite.structure.io.pdbx as pdbx
import biotite.database.rcsb as rcsb
import ammolite
PNG_SIZE = (800, 800)
assembly = pdbx.get_assembly(
pdbx.PDBxFile.read(rcsb.fetch("1XI4", "cif")),
model=1
)
# Structure contains only CA
# Bonds are not required for visulization -> empty bond list
assembly.bonds = struc.BondList(assembly.array_length())
# General configuration
ammolite.cmd.bg_color("white")
ammolite.cmd.set("cartoon_side_chain_helper", 1)
ammolite.cmd.set("cartoon_oval_length", 0.8)
ammolite.cmd.set("depth_cue", 0)
ammolite.cmd.set("valence", 0)
pymol_obj = ammolite.PyMOLObject.from_structure(assembly)
pymol_obj.show_as("spheres")
ammolite.cmd.set("sphere_scale", 1.5)
ammolite.show(PNG_SIZE)

chain_ids = np.unique(assembly.chain_id)
for chain_id, color in zip(chain_ids, plt.get_cmap("tab20").colors):
pymol_obj.color(color, assembly.chain_id == chain_id)
ammolite.show(PNG_SIZE)
# sphinx_gallery_thumbnail_number = 2